
- Trainer/in: Arthur Brommer
- Trainer/in: Thomas Cremer
- Trainer/in: Wolfgang Enard
- Trainer/in: Ines Hellmann
- Trainer/in: Eva Briem
- Trainer/in: Johanna Geuder
- Trainer/in: Ines Hellmann
- Trainer/in: Manqi Liang
- Trainer/in: Daniel Richter
- Trainer/in: Johanna Geuder
- Trainer/in: Ines Hellmann
- Trainer/in: Daniel Richter
- Trainer/in: Lucas Wange
- Trainer/in: Marion Cremer
- Trainer/in: Thomas Cremer
- Trainer/in: Wolfgang Enard
- Trainer/in: Benedikt Grothe
- Trainer/in: Stephan Sellmaier
- Trainer/in: Cora Stuhrmann

- Trainer/in: Wolfgang Enard
- Trainer/in: Ines Hellmann
Content: This lecture builds on knowledge obtained in molecular biology and genetics on the Bachelor's level. It aims to deepen an understanding how the human genome was sequenced and annotated and how it is currently used to study human biology in health and disease. The following topics are addressed: The human genome project, high throughput sequencing technologies, basics in sequence analysis, gene annotation, repeats, gene expression analysis.
Qualification goals: The students will be able to describe and understand fundamental principles of human genomic research. They will acquire the basic background knowledge to apply genomic technologies.

- Trainer/in: Wolfgang Enard
- Trainer/in: Ines Hellmann
- Trainer/in: Daniel Richter

- Trainer/in: Julia Bechteler
- Trainer/in: Cordelia Bolle
- Trainer/in: Elsbeth Bösl
- Trainer/in: Laura Busse
- Trainer/in: Wolfgang Enard
- Trainer/in: Marc Gottschling
- Trainer/in: Annika Guse
- Trainer/in: Ines Hellmann
- Trainer/in: Martin Heß
- Trainer/in: Kirsten Jung
- Trainer/in: Gudrun Kadereit
- Trainer/in: Hans-Henning Kunz
- Trainer/in: Dario Leister
- Trainer/in: Heinrich Leonhardt
- Trainer/in: Daniela Meilinger
- Trainer/in: Jörg Meurer
- Trainer/in: Birgit Neuhaus
- Trainer/in: Kärin Nickelsen
- Trainer/in: Reinhard Obst
- Trainer/in: Martin Parniske
- Trainer/in: Herwig Stibor
- Trainer/in: Cora Stuhrmann
- Trainer/in: Marie Veranso Epse Libalah
- Trainer/in: Grazyna Wlodarska-Lauer
- Trainer/in: Jochen Wolf
- Trainer/in: Ming Zhao
- Trainer/in: Albert Zink
Computational early drug discovery in cancer; Analysing drug high-throughput screens of cancer cell lines and their deep molecular characterisation for biomarker discovery; Investigating putative drug targets with CRISPR depletion screens. Retrieving insights in the disease aetiology of cancer by analysing large patient cohorts; In this purely computational course, we will investigate pharmacogenomic data, which will be harnessed by applying biostatistical and machine learning methods.
- Trainer/in: Alina Arneth
- Trainer/in: Gülce Gökçe
- Trainer/in: Ines Hellmann
- Trainer/in: Linus Hölzel
- Trainer/in: Michael Menden
- Trainer/in: Matthias Meyer-Bender
- Trainer/in: Barbara Streibl
Current topics in our research field are discussed and priority is given for for students doing research course, Bachelor thesis or Master thesis; This seminar is only recommended for advanced students with an aptitude for quantitative and statistical approaches. 3 ECTS Points
Content: In the seminar, the students critically present and discuss current publications related to genomic analyses.
Qualification goals: The students will be able to extract and judge relevant information also from complex literature and to exchange information and ideas on a scientific level with experts in Genomics.
- Trainer/in: Wolfgang Enard
- Trainer/in: Ines Hellmann
- Trainer/in: Aleksandar Janjic
- Trainer/in: Philipp Janßen
- Trainer/in: Beate Vieth

- Trainer/in: Wolfgang Enard
- Trainer/in: Ines Hellmann
- Trainer/in: Daniel Richter
- Trainer/in: Pascal Stümpfl
Handling statistical data is the key to success in many fields of biology. In this practical course we will
- discuss how you can use plots to make friends with your data
- discuss how you prepare publication ready plots
- repeat some basic concepts in of statistics
- apply the concepts to interpret published figures
All this will be accomplished using R and ggplot2.
The course consists of 2 parts:
1. Practical part (3ECTS, final exam is a term paper)
2. Seminar ( 3ECTS, prepare & present a plot that illustrates a data-centric question of your choosing e.g. about COVID-19 stats)
The enrolement key is: PrettyPlots
- Trainer/in: Ines Hellmann
- Trainer/in: Dana Lopez Parra
- Trainer/in: Felix Pförtner
- Trainer/in: Ines Hellmann
- Trainer/in: Dana Lopez Parra
- Trainer/in: Felix Pförtner
- Trainer/in: Daniel Richter
- Trainer/in: Anita Térmeg
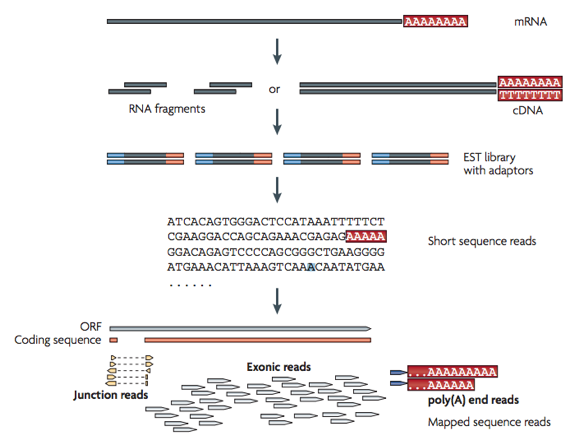
This seminar introduces methods for the computational analysis of quantitative high-throughput RNA sequencing data (RNA-Seq). The seminar is required for the practical course "Computational analysis of RNA-Seq" and the lecture "Human Genomics" is highly recommended.
Instructors: Ines Hellmann hellmann@bio.lmu.de
Beate Vieth vieth@bio.lmu.de
Please remember that you have to consult with one of us at least 2 days prior to the Seminar.
Talks should be 20min/person and including discussion each section should be roughly 1.5h.
The points given in each topics are hints, you may restructure it and make slight changes to the emphasis if you think it to be appropriate.
Talks are generally better if, you consider the following principles:
Less text, more pictures
Text and pictures need to be in sync
Explain everything to a level that your fellow students understand the pictures and statements you make. Don't just show things for the sake showing.
- Trainer/in: Ines Hellmann
- Trainer/in: Philipp Janßen
- Trainer/in: Zane Kliesmete
- Trainer/in: Dana Lopez Parra
- Trainer/in: Felix Pförtner
- Trainer/in: Daniel Richter
- Trainer/in: Leonhard Schaffmayer
- Trainer/in: Anita Térmeg